The Challenge of Specific Cathepsin S Activity Detection in Experimental Settings
Alex Steimle and Julia-Stefanie Frick*
Abstract
In recent years, a growing interest in pathophysiological processes that are associated with the endosomal and lysosomal protease cathepsin S (CTSS) results in an increasing number of various published methods for CTSS activity detection. CTSS has been reported to be involved in the pathology of autoimmune diseases like multiple sclerosis as well as in tumor growth and Alzheimer’s disease. These implications make this enzyme a first class drug target. In order to fully understand the involvement of CTSS in the formation of pathological processes, gene and protein expression analysis is not sufficient. Rather, one should focus on the regulation of its enzymatic activity. Different approaches for CTSS activity detection are available and described. However, some of these approaches are not suitable for a standard laboratory without special equipment or technical expertise or provide other limitations. We have recently published an easy-to-perform protocol for reliable, quantifiable and reproducible CTSS activity detection. In this review we want to discuss our application and compare it with other published methods and protocols. This might help researchers who are interested in CTSS research to decide which application fits best to their technical or personal facilities.
Text
Cathepsin S (CTSS) is a protease located in lysosomes or endosomes of professional antigen presenting cells (APC), such as macrophages, dendritic cells and B cells1. Dysregulated CTSS expression and/or activity has been reported to be involved in the pathogenesis of various diseases. CTSS is the major regulator of major histocompatibility complex (MHC) II surface expression. Therefore, especially autoimmune diseases which are caused by (or which are associated with) pathologically enhanced CD4+ T cell activation are part of this disease portfolio. In this context, the Sjögren’s syndrome2, atherosclerosis3, psoriasis4 and an animal model of rheumatic arthritis5 play dominant roles. But not only autoimmune diseases are associated with CTSS dysregulation. Enhanced CTSS activity could also be detected in the bronchoalveolar lavage of cystic fibrosis patients6. Neurologists might focus on the role of CTSS in the development of multiple sclerosis (MS) and experimental autoimmune encephalomyelitis (EAE), the corresponding mouse model of MS7. Indeed, it could already been shown that CTSS is capable of myelin basic protein proteolysis8 which might contribute to the observed phenotypes in MS patients. In fact, enhanced CTSS levels could be found in peripheral blood mononuclear cells, serum and the cerebrospinal fluid of MS patients9-12. In line with these observations, chemical inhibition of CTSS led to a reduction of phenotypical severity in the EAE mouse model5,13. Besides MS patients, enhanced CTSS activity at the primary site in breast cancer patients correlated with an increased frequency of the appearance of brain metastases14. The implication of CTSS concerning cancer was linked with with tumor-associated APCs, namely macrophages15. Alzheimer’s disease (AD) was already reported to be associated with a malfunctioning Cathepsin B (CTSB) activity regulation16,17. But also human Cathepsin S seems to be involved in the manifestation of the disease phenotype since also CTSS provides β-secretase activity18which leads to the AD phenotype causing agent Aβ peptide through amyloid precursor protein proteolysis19. One important feature of CTSS, distinguishing it from other cysteine cathepsins, is the retention of its proteolytic activity at neutral pH. Being part of the endosomal and lysosomal compartment, CTSS can be secreted into the extracellular space providing neutral pH where it can cause additional pathologies. Extracellular CTSS activity has already been shown to be involved in activating protease-activated receptors (PAR) like PAR2 as well as G-protein coupled receptors like Mrgprs leading to the promotion of itch and pain sensation in both cases20-23.
All the above mentioned implications for CTSS activity dysregulation which are linked with manifestation of various pathologies makes this enzyme an attractive and intensely studied drug target. Some studies just refer to enhanced Ctss gene expression rates or increased protein amounts inside or outside of target cells. However, mRNA expression levels of a protease-encoding gene or expression of the corresponding protein does not necessarily correlate with the overall detected activity of the respective enzyme. This is due to several possible posttranscriptional regulation mechanisms that can occur inside or outside a eukaryotic cell. Proteases can be (i) translated as inactive zymogens requiring proteolytic activation, (ii) activated by co-factors, (iii) separated from their substrates by localization in distinct intra- or extracellular compartments or (iv) inhibited by binding of endogenous inhibitors to the active site of the protease24. All these examples illustrate that focusing on mRNA or protein expression is not sufficient to reliably investigate the role of proteases in biological or pathophysiological processes. Therefore, specific detection of the activity of a certain protease has to be guaranteed in order to exclude false positive detection of one or more other proteases with a similar substrate specifity. This is a grave problem concerning the protease group “cathepsins”, which consists of currently eleven known members25, since cathepsins in general provide a broad and overall similar substrate specifity26. This makes CTSS activity detection a challenge, in vitro, ex vivo and especially in vivo.
But how can an experimentator achieve the goal to reliably detect CTSS activity in the biological system of choice? There are several possibilities how CTSS activity can be detected, all of them with their characteristic advantages and limitations. A suitable CTSS activity detection method should match the following four criteria: (i) high specifity, (ii) high sensitivity since CTSS expression is restricted to APC, (iii) the experimental workflow should be able to be performed in a more or less standard laboratory without the requirement of highly special technical equipment and (iv) the method should deliver reproducible and quantifiable results. We summarize some available techniques for CTSS activity detection with their characteristic pros and cons in table 1.
Popular approaches include activity based probes (ABPs). These are small reporters of proteolytic activity which bind covalently and irreversibly to the active site of the target enzymes27 leading to a loss of enzymatic activity. This irreversible CTSS inhibition should be kept in mind if in vivo monitoring shall be performed. ABPs usually consist of three distinct functionalities (Fig. 1A): (1) The so-called warhead functionality leads the probe to enzymes sharing a common catalytic mechanism, (2) the recognition element that enhances specificity for one or more specific enzymes and (3) a tag for a later detection of the tagged enzymes. Such ABPs exist for the detection of human and mouse CTSS. General disadvantages of such ABPs include a lack of specificity for CTSS for some of these ABPs. This has then to be compensated by further time-consuming experimental approaches like western blotting, immunoprecipitations etc. Additionally, CTSS activity can often only be determined in a semi-quantitative manner like measuring band intensities on a gel or a detection of overall fluorescence intensity. However, the potential labelling of proteolytically active CTSS in live cells due to cell permeability is a big advantage of some ABPs. Veilleux et al. developed radioiodinated ABPs to selectively label active CTSS in human whole blood28. Ben-Aderet et al.29 and Oresic Bender et al.30 used fluorogenic ABPs to monitor enzymatically active CTSS in human cells or live mice, respectively. Other examples of recent ABPs development for CTSS activity detection include the publications of Barlow et al.31, Hughes et al.32, Mertens et al.33 and Garenne et al34. However, some of them provide a lack of specificity for CTSS and also label other cathepsins29,32-35 while other ABPs selectively label human or mouse CTSS28,30,36. Another general disadvantage of ABPs include the requirement of more specialized detection techniques that a non-specialized lab could tentatively not afford.
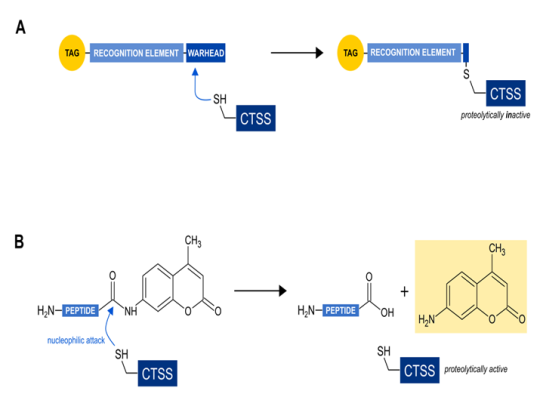
Figure 1: Functional comparison of the two major classes of probes for direct CTSS activity detection
(a) activity based probes (ABPs): The so-called warhead covalently links the probe with CTSS, leading to the inhibition of its enzymatic activity. The recognition element supports the specificity for CTSS alone or some related cysteine cathepsins. The tag allows for later detection of the tagged enzymes.
(b) substrate based probes (SBPs): The thiol group at the active site of CTSS nucleophilically attacks the peptide bond of the peptidyl substrate leading to a deliberation of the aminomethylcoumarin. The resulting change in the fluorogenic behaviour of free aminomethylcoumarin can then be detected in a fluorescent compatible photometer.
Recently, commercially available CTSS activity detection kits have been developed and are sold. Experiences made by our group revealed, however, that these kits are far less specific than they are advertised to be and we strongly recommend to use alternative techniques for reliable CTSS activity detection. If this is not possible, a strict validation of the claimed specificity for CTSS should be performed, i.e. using recombinant enzymes or knock-out cells for various cysteine cathepsins.
Other approaches include an indirect method to determine CTSS proteolytic (table 1) activity by not directly assessing the enzymatic activity but rather determining the intracellular amount of the MHC II-bound invariant chain fragment (Iip10), which is a substrate for CTSS. Therefore, decreased amounts of Iip10 is stated to correlate with enhanced CTSS activity5. Since CTSS is translated into an inactive pro-enzyme requiring proteolytical cleaving which results in a 24 kDa active enzyme, some publications refer to the band intensities of this 24 kDa band when making statements on CTSS activity37. However this does not take the potential binding of endogenous inhibitors into account.
| Application category | Characteristic | Example | target | Year published | Quantification method | Limitation | Outstanding advantage |
|---|---|---|---|---|---|---|---|
| Activity based probe (ABPs) | covalent binding to CTSS active site leading to enzyme inhibition | GB12329 | Human CTSS and CTSB | 2015 | Fluorescent label | Binds also to CTSB. Gel-assisted enzyme separation or IP needed | Non invasive in vivo imaging possible |
| Probe 736 | Human CTSS | 2015 | Fluorescent label | Not cell permeable | Specificity for CTSS | ||
| Z-PraVG-DMG32 | Human CTSS, CTSB and CTSL | 2016 | Rhodamineazide labelling | Similar Ki values for CTSS, CTSB and CTSL | Cell-permeability of the substrate | ||
| CM-27935 | Human CTSS and CTSL | 2013 | Luciferase assisted light detection | Also labels CTSL | Reliable detection of combined CTSS and CTSL activity | ||
| BMV15730 | Mouse CTSS | 2015 | Fluorescent label | availability | Highly specific, in vivo and in vitro application possible | ||
| BIL-DMK28 | Human CTSS | 2011 | Radioactive label | Radioactivity | Specificity for CTSS | ||
| Probe 1033 | Human CTSS, CTSL, CTSB and CTSK | 2014 | Fluorescent label | Not specific for CTSS | Suitable for complex protein mixtures | ||
| Biot-(PEG)2-Ahx-LeuValGly-DMK34 | Human CTSS, CTSL, CTSB and CTSL | 2015 | SDS Page assisted chemi-luminescence | Not specific for CTSS | Easy to perform | ||
| Substrate based probes (SBP) | Substrate turnover; enzyme is not inhibited and remains proteolytically active | PMGLP | Human CTSS43 and mouse CTSS42 | 2016 | Fluorescence increase | Requires cell lysis and inhibition control | Simple enzymatic assay, easy-to-perform, time saving. |
| Ac-KQKLR-AMC | Mouse CTSS | 2014 | Fluorescence increase | Requires cell lysis and inhibition control | Simple enzymatic assay | ||
| NB20031 | Mouse CTSS, CTSK and CTSB | 2015 | Fluorescence increase | Detects also CTSK at pH7.5 and CTSB at pH 5.5 | Simple enzymatic assay | ||
| Z-FR-AMC | Mouse CTSS, CTSB and CTSL | 2002 | Fluorescence increase | Unspecificity, requires extensive experimental setting | Simple enzymatic assay | ||
| Indirect detection methods | Does not directly interact with CTSS | Detection of CTSS substrate accumulation by western blotting3 | Mouse invariant chain p10 detection | 2015 | Western blot band intensity of Iip10 | Does not detect CTSS activity directly | Simple to perform |
| Western blotting of 24 kDa CTSS37 | Human CTSS | 2014 | Western blot band intensity of 24 kDa CTSS | Does not consider potential binding of endogenous inhibitors and does not refer to activity | Simple to perform |
Table 1. Summary of various CTSS activity detection techniques. The techniques are grouped into the major functional classes
Another possibility for direct CTSS activity determination is to use substrate-based probes (SBP) like i.e. fluorochrome-coupled peptidyl substrates. The principle mechanism includes a detection in the increase of a peptidyl-coupled and later proteolytically liberated fluorochrome (Figure 1B). SBPs provide the advantage of not inhibiting the target enzyme. Prominent examples are Z-VVR-AMC38, Z-FR-AMC36,39 or Ac-KQKLR-AMC20,40. All of them share the aminomethyl coumarin group as a fluorogen tag. Coumarins are popular fluorescent labels due to their large stoke’s shifts41 and their small size allowing for incorporation into small peptides33. These peptidyl substrates generally provide the disadvantage to be usable only in cell lysates and not in living cells or organisms. The major potential problem with most peptidyl substrates is the unsatisfactory specificity for CTSS. Due to the already mentioned similar substrate specificity among cysteine cathepsins, other cathepsins than CTSS can deliver false positive results due to unspecific cleavage of the substrate. However, we have recently reported on a CTSS activity detection method using a coumarin-coupled peptidyl substrate (PMGLP) that allows for highly specific CTSS activity detection in mouse samples42. This method is a simple standard enzymatic assay detecting a time-dependent increase in fluorescence intensity caused by cleaving of the peptidyl sequence. It is easy-to-perform, time-saving and can be used for high-throughput applications. Additionally, a standard curve using free aminomethyl coumarin can be created allowing for the detection of specific enzymatic activities within a tested sample which is a big advantage over ABPs. The oligopeptidyl substrate we use was initially published as being suitable for highly specific detection of CTSS activity in human antigen presenting cells43. However, we could demonstrate that is also suitable for mouse samples. The substrate, Mca-GRWPPMGLPWE-Lys(Dnp)-DArg-NH2 (PMGLP) was shown to be cleaved specifically by murine CTSS at a pH of 7.5 with no other murine cathepsin contributing to substrate cleaving in cell lysates42. The obtained results were easily quantifiable and highly reproducible as demonstrated in mouse bone marrow derived dendritic cells that were differentiated from the bone marrow of different individuals. The presented method therefore delivers specific CTSS activities and only requires a phosphate buffer system and a fluorescence compatible heatable photometer making the application attractive for less specialized laboratories. We recommend the following simple workflow (Figure 2): the target cells are lysed hypotonically with the support of freezing in liquid nitrogen. This guarantees a minimal loss of CTSS activity due to cell lysis. Protein concentrations of the lysates are determined afterwards and each sample is separated into two samples. The first one contains the substrate, PMGLP, and the second one PMGLP and E-64, a cysteine cathepsin inhibitor that is easily commercially available and cheap. The latter sample is used as a negative control. Substrate conversion is detected for at least 20 min at 37°C by detection of fluorescence increase at 405 nm after excitation at 340 nm. Conversion rates are computed by determination of the slopes of the linear fluorescence increase and the slope of the inhibitor treated negative control is substracted from the slope of the non-inhibitor treated sample. This workflow is fast and the obtained results are highly specific for murine CTSS as demonstrated in our published protocol42. We therefore hope to contribute to a progress in CTSS related research.
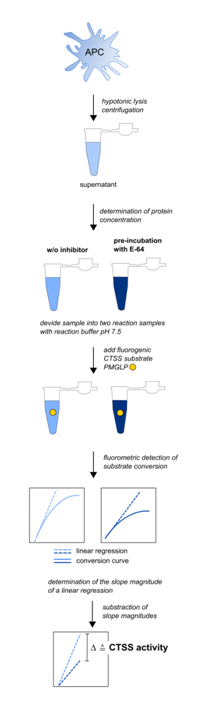
Figure 2: Experimental workflow for the detection of CTSS activities in mouse APC (adapted from Steimle et al., 2016)
Cells of interest are lysed hypotonically followed by blast freezing in liquid nitrogen as described in materials and methods section. Supernatant is used for CTSS activity detection. Out of each sample, two reaction samples containing the same amount of whole cell protein are created. The first sample contains the fluorogen substrate, the second both, the substrate and the cathepsin inhibitor E-64. Reaction buffer provides a pH of 7.5 to minimize false positive substrate conversion caused by other cathepsins. Substrate conversion is monitored for 60 min at 37 °C. A linear regression at the very beginning of the sigmoidal conversion curve is performed for both reaction samples. The slope magnitude of this linear regression is equivalent to the substrate conversion per time unit. The difference in the slope magnitude of both reaction samples is equivalent to the CTSS activity in the sample the lysate was taken from. CTSS activities can therefore be expressed as the difference of fluorescence intensity per µg cell protein and time unit, i.e. ?AU min-1 µg-1.
Acknowledgements
Work was supported by the DFG (DFG FR 2087/6-1, DFG FR 2087/8-1, CRC685, SPP1656), the Bundesministerium für Bildung and Forschung (BMBF) and the German Centre for Infection Research (DZIF)
References
- Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, Rudensky AY. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol. 2005;174(3):1205-12.
- Hamm-alvarez SF, Janga SR, Edman MC, et al. Tear cathepsin S as a candidate biomarker for Sjögren's syndrome. Arthritis Rheumatol. 2014;66(7):1872-81.
- Figueiredo JL, Aikawa M, Zheng C, et al. Selective cathepsin S inhibition attenuates atherosclerosis in apolipoprotein E-deficient mice with chronic renal disease. Am J Pathol. 2015;185(4):1156-66.
- Schönefuss A, Wendt W, Schattling B, et al. Upregulation of cathepsin S in psoriatic keratinocytes. Exp Dermatol. 2010;19(8):e80-8.
- Baugh M, Black D, Westwood P, et al. Therapeutic dosing of an orally active, selective cathepsin S inhibitor suppresses disease in models of autoimmunity. J Autoimmun. 2011;36(3-4):201-9.
- Weldon S, Mcnally P, Mcauley DF, et al. miR-31 dysregulation in cystic fibrosis airways contributes to increased pulmonary cathepsin S production. Am J Respir Crit Care Med. 2014;190(2):165-74.
- Baxter AG. The origin and application of experimental autoimmune encephalomyelitis. Nat Rev Immunol. 2007;7(11):904-12.
- Beck H, Schwarz G, Schröter CJ, et al. Cathepsin S and an asparagine-specific endoprotease dominate the proteolytic processing of human myelin basic protein in vitro. Eur J Immunol. 2001;31(12):3726-36.
- Nagai A, Murakawa Y, Terashima M, et al. Cystatin C and cathepsin B in CSF from patients with inflammatory neurologic diseases. Neurology. 2000;55(12):1828-32.
- Staun-ram E, Miller A. Cathepsins (S and B) and their inhibitor Cystatin C in immune cells: modulation by interferon-β and role played in cell migration. J Neuroimmunol. 2011;232(1-2):200-6.
- Bever CT, Garver DW. Increased cathepsin B activity in multiple sclerosis brain. J Neurol Sci. 1995;131(1):71-3.
- Haves-zburof D, Paperna T, Gour-lavie A, Mandel I, Glass-marmor L, Miller A. Cathepsins and their endogenous inhibitors cystatins: expression and modulation in multiple sclerosis. J Cell Mol Med. 2011;15(11):2421-9.
- Fissolo N, Kraus M, Reich M, et al. Dual inhibition of proteasomal and lysosomal proteolysis ameliorates autoimmune central nervous system inflammation. Eur J Immunol. 2008;38(9):2401-11./li>
- Sevenich L, Bowman RL, Mason SD, et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat Cell Biol. 2014;16(9):876-88.
- Verdoes M, Edgington LE, Scheeren FA, et al. A nonpeptidic cathepsin S activity-based probe for noninvasive optical imaging of tumor-associated macrophages. Chem Biol. 2012;19(5):619-28.
- Hook V, Toneff T, Bogyo M, et al. Inhibition of cathepsin B reduces beta-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate beta-secretase of Alzheimer's disease. Biol Chem. 2005;386(9):931-40.
- Hook V, Schechter I, Demuth HU, Hook G. Alternative pathways for production of beta-amyloid peptides of Alzheimer's disease. Biol Chem. 2008;389(8):993-1006.
- Schechter I, Ziv E. Cathepsins S, B and L with aminopeptidases display β-secretase activity associated with the pathogenesis of Alzheimer's disease. Biol Chem. 2011;392(6):555-69.
- Poeck B, Strauss R, Kretzschmar D. Analysis of amyloid precursor protein function in Drosophila melanogaster. Exp Brain Res. 2012;217(3-4):413-21.
- Zhao P, Lieu T, Barlow N, et al. Cathepsin S causes inflammatory pain via biased agonism of PAR2 and TRPV4. J Biol Chem. 2014;289(39):27215-34.
- Cattaruzza F, Lyo V, Jones E, et al. Cathepsin S is activated during colitis and causes visceral hyperalgesia by a PAR2-dependent mechanism in mice. Gastroenterology. 2011;141(5):1864-74.e1-3.
- Reddy VB, Sun S, Azimi E, Elmariah SB, Dong X, Lerner EA. Redefining the concept of protease-activated receptors: cathepsin S evokes itch via activation of Mrgprs. Nat Commun. 2015;6:7864.
- Reddy VB, Shimada SG, Sikand P, Lamotte RH, Lerner EA. Cathepsin S elicits itch and signals via protease-activated receptors. J Invest Dermatol. 2010;130(5):1468-70.
- Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5(9):785-99.
- Turk V, Stoka V, Vasiljeva O, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824(1):68-88.
- Choe Y, Leonetti F, Greenbaum DC, et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281(18):12824-32.
- Serim S, Haedke U, Verhelst SH. Activity-based probes for the study of proteases: recent advances and developments. ChemMedChem. 2012;7(7):1146-59.
- Veilleux A, Black WC, Gauthier JY, et al. Probing cathepsin S activity in whole blood by the activity-based probe BIL-DMK: cellular distribution in human leukocyte populations and evidence of diurnal modulation. Anal Biochem. 2011;411(1):43-9.
- Ben-aderet L, Merquiol E, Fahham D, et al. Detecting cathepsin activity in human osteoarthritis via activity-based probes. Arthritis Res Ther. 2015;17:69.
- Oresic bender K, Ofori L, Van der linden WA, et al. Design of a highly selective quenched activity-based probe and its application in dual color imaging studies of cathepsin S activity localization. J Am Chem Soc. 2015;137(14):4771-7.
- Barlow N, Nasser Y, Zhao P, et al. Demonstration of elevated levels of active cathepsin S in dextran sulfate sodium colitis using a new activatable probe. Neurogastroenterol Motil. 2015;27(11):1675-80.
- Hughes CS, Shaw G, Burden RE, Scott CJ, Gilmore BF. The application of a novel, cell permeable activity-based probe for the detection of cysteine cathepsins. Biochem Biophys Res Commun. 2016;472(3):444-50.
- Mertens MD, Schmitz J, Horn M, et al. A coumarin-labeled vinyl sulfone as tripeptidomimetic activity-based probe for cysteine cathepsins. Chembiochem. 2014;15(7):955-9.
- Garenne T, Saidi A, Gilmore BF, et al. Active site labeling of cysteine cathepsins by a straightforward diazomethylketone probe derived from the N-terminus of human cystatin C. Biochem Biophys Res Commun. 2015;460(2):250-4.
- Cagli? D, Repnik U, Jedeszko C, et al. The proinflammatory cytokines interleukin-1α and tumor necrosis factor α promote the expression and secretion of proteolytically active cathepsin S from human chondrocytes. Biol Chem. 2013;394(2):307-16.
- Kohl F, Schmitz J, Furtmann N, et al. Design, characterization and cellular uptake studies of fluorescence-labeled prototypic cathepsin inhibitors. Org Biomol Chem. 2015;13(41):10310-23.
- Castellano J, Badimon L, Llorente-cortés V. Amyloid-β increases metallo- and cysteine protease activities in human macrophages. J Vasc Res. 2014;51(1):58-67.
- Magister S, Obermajer N, Mirkovi? B, et al. Regulation of cathepsins S and L by cystatin F during maturation of dendritic cells. Eur J Cell Biol. 2012;91(5):391-401.
- Schwarz G, Boehncke WH, Braun M, et al. Cathepsin S activity is detectable in human keratinocytes and is selectively upregulated upon stimulation with interferon-gamma. J Invest Dermatol. 2002;119(1):44-9.
- Balce DR, Allan ER, Mckenna N, Yates RM. γ-Interferon-inducible lysosomal thiol reductase (GILT) maintains phagosomal proteolysis in alternatively activated macrophages. J Biol Chem. 2014;289(46):31891-904.
- Katritzky AR, Cusido J, Narindoshvili T. Monosaccharide-based water-soluble fluorescent tags. Bioconjug Chem. 2008;19(7):1471-5.
- Steimle A, Kalbacher H, Maurer A, et al. A novel approach for reliable detection of cathepsin S activities in mouse antigen presenting cells. J Immunol Methods. 2016;432:87-94.
- Lützner N, Kalbacher H. Quantifying cathepsin S activity in antigen presenting cells using a novel specific substrate. J Biol Chem. 2008;283(52):36185-94.
