The Importance of Data Sources for Machine Learning Applications in Autism: A Mini Review
Itzhak Kurek*, Jean-Christophe Quillet, Michael Siani-Rose
Cannformatics, Inc., San Francisco, CA, USA
Abstract
Autism spectrum disorder (ASD) is a group of lifelong heterogeneous neurodevelopmental conditions with a wide range of severity levels that affect social communication and social interaction. Diagnosis of ASD relies on subjective observation of these clinical phenotypes. The growing body of big data generated by subjective methods and more recently by objective high-throughput technologies such as omics for the detection of biomolecules, is being successfully applied to a rapidly-growing number of machine learning (ML) algorithms to inform research for diagnostics and interventions for patients with ASD. While most reviews in this area are focused on the ML approaches, we highlight the impact of the database on the expected outcomes in ML-based ASD research studies.
Introduction
Autism spectrum disorder (ASD) is a set of neurodevelopmental conditions diagnosed by a qualified clinician such as a developmental pediatrician or neurologist1. It is characterized by qualitative impairments in social interaction and communication, as well as restricted, repetitive, and/or stereotyped patterns of behavior2. ASD diagnosis is not a straightforward process and is often made long after initiation. In most cases, assessment is reliable at the age of 2 years3, and sometimes at 18 months4, while onset can occur as early as the first- or second-trimester5 as fever-associated immune disturbances in response to prenatal infectious agent exposure6 lead to a pleiotropic effect on metabolic pathways7.
There is no “one-pill-fits-all” approach for ASD treatment. Personalized educational and behavioral therapies are the main approaches, supplemented with prescription medication in 48% of children8. Evaluation of treatment effectiveness in children with ASD is challenging due to the variability in symptoms expressed and in severity levels both among children with ASD and within each child over time. Also, it requires stepwise assessments that often involve family member and care giver interactions. Dykens et al.9 showed that these in-person interactions for ASD evaluation can introduce additional variation resulting from the child’s distress, impacting the outcome.
The significant increase in the availability of data and machine learning (ML) algorithms presents new opportunities to diagnose, distinguish categories of patients, predict and monitor the efficacy of therapy, and identify the underlying conditions of ASD. ML is an artificial intelligence (AI) branch based on algorithms and statistical models that learn and improve from experience without being explicitly programmed by drawing inferences from patterns in data to make predictions or decisions based on those patterns.
This minireview aims to provide an overview of ASD ML studies with a focus on the importance of database selection for ML applications and the ability to incorporate bioinformatics tools such as systems biology and disease genomics to achieve the desired outcomes.
Data Sources
Significant increases in the number of ASD cases, the amount of ASD-related data from multiple technologies (e.g. genomics, rs-fMRI, etc.) and the number of sources (e.g. national databases, foundations for ASD research etc.) are currently driving the growth in database sources available for ML applications10,11. These sources include data resources with phenotypic and genetic data of thousands to tens of thousands of participants per database such as the National Database for Autism Research (NDAR), Simons Foundation Autism Research Initiative (SFARI), and the Autism Genetic Resource Exchange (AGRE).
However, ML predictions in life sciences are heavily dependent on high quality data characterized by: 1) correct experiment design - investigators can estimate the errors and understand the bias and sensitivity of the data; 2) standardization of data repositories - the processes for data extraction, analysis, and quality control are standardized; and 3) reproducibility - statistical design and analysis that ensure reproducibility of a study at the experimental, empirical, computational and ethical levels12. While the information regarding experimental design and data repositories are documented in the source, reproducibility is often an unknown factor, with higher impact in data from subjective evaluation. In a study aiming to determine the validity of the findings of 100 peer-reviewed studies published in three psychology journals, the authors found 50% of the studies could not be reproduced13. Although successful replication provides only validity of the results, it is a prerequisite for medical and physiological interpretation of ML predictions.
The availability of multiple high-quality data sources for clinical phenotypes obtained via a variety of modalities including observations by parents, clinicians, video, and audio devices11 and omics techniques that numerically quantify fundamental biological processes14 have the potential to associate behavior with omics information in children with ASD, but present challenges for interpretation. For example, linking available data from clinical phenotypes of ASD to genetic factors such as the high-conï¬dence ASD (hcASD) genes during fetal development5, environmental factors such as maternal nutrition, viral and bacterial infections15, and cultural beliefs at the community-level that can delay early intervention and impact the severity clinical phenotypes of ASD16, is possible, but interpretation is difficult.
Figure 1 illustrates the general path from genotype to clinical phenotype and is divided into two parts: 1) the cellular, which is evaluated by objective omics methods; and 2) the clinical phenotype, in which subjective human interpretation is required during the process. Although the figure presents a straight line between the two parts, results are not entirely a continuum, as objective and subjective evaluations capture different aspects of the diagnosis and act as complementing rather than overlapping information. This can affect the ability of ML to predict clinical phenotype directly from genetics.
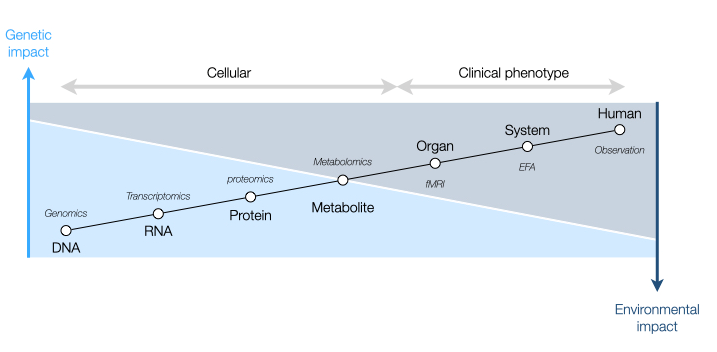
Figure 1: A simplified presentation of the hierarchical path from genotype to clinical phenotype with respect to the effect of genetics and environment in patients with ASD and the indicated methods to extract data and integrate it into a dataset.
Cellular Level
At the single-cell level, the technologies to extract data belong to the omics disciplines, a suffix used in life sciences to describe the large-scale data/ information required to understand a complete biological system17. Using cellular features such as DNA and mRNA in a high-throughput manner, researchers can characterize different biological systems in a static or dynamic mode and connect the information from DNA all the way downstream to a metabolite. Genomics databases can integrate with disease genomics to identify disease-associated genes and disease-causing mutation biomarkers, and multiple omics databases can integrate with bioinformatics platforms such as systems biology to construct networks, predict interactions and monitor dynamic responses18. Since gene expression is regulated at the mRNA and protein levels from transcription initiation to protein degradation, metabolomics has the best tools to link the individual physiological/pathophysiological state to both downstream objective methods and upstream subjective methods while factoring in the impact of genetics, environmental stimuli, diet, and gut microbiome19.
System Level
At the system level (Figure 1), upstream to the omics disciplines is the brain image-derived phenotype, a quantifiable data-driven approach that associates brain activity and clinical phenotype of ASD. Linked to a specific area in the brain, functional magnetic resonance imaging (fMRI) is a noninvasive functional imaging technique in which the metabolic activity of tissues is determined indirectly via oxygen consumption20. The resting-state fMRI (rs-fMRI) is an advanced alternative that quantifies the spontaneous brain activity of an individual in the absence of stimuli (during resting).
Subjective data sources from modalities such as eye gaze and atypical ASD-specific motor phenotype of slow responses for finger tapping can be quantified by eye movement and kinematic tests21. The gold standard for ASD diagnosis is the complementary Autism Diagnostic Observation Schedule (ADOS) and the Autism Diagnostic Interview – Revised (ADI-R), the main data sources, together with the Childhood Autism Rating Scale (CARS) and Gilliam Autism Rating Scale (GARS)11. Parental multiple-choice questionnaires22 and Likert scale surveys23 are often used with other data sources in ASD studies.
Linking omics technology and behavioral assessment was previously reported by Bent et al.24 in a study that statistically correlated clinical phenotype in children with ASD treated with a sulforaphane supplement from broccoli and metabolomics. In this study, parental reports suggested a metabolic link between sphingolipids/ sphingomyelins and improvement in clinical phenotype. A recent study by Quillet et al.25 identified biomarkers distinguishing ASD and typically developing (TD) groups and linked the cannabinoids tetrahydrocannabinol (THC), cannabidiol (CBD) and cannabigerol (CBG) with metabolite levels in children with ASD. It was the first to use ML algorithms on a pharmacometabolomics dataset of previously identified cannabis-responsive biomarkers and other metabolites in children with ASD that shift toward physiological levels determined in typically developing children (TD) after successful medical cannabis treatment23,26.
Machine Learning Applications
Since 2012, researchers have trained ML algorithms on a wide range of data types to improve diagnostic processes and the understanding of ASD11,27. ML applications are often used to facilitate the direct diagnosis of ASD in individual patients, integrate observational data, and facilitate the analysis of parent-reported questionnaires and reported behavior from home-recorded videos22, and/or kinematic and motion features from video recordings of adults21. ML applications are also used for ASD biomarker discovery, training on data acquired from a broad range of technologies: fMRI20, metabolomics25,28, proteomics29, transcriptomics30 or a combination of those31. At the genome level, ML has been applied for functional characterization of the genetic basis of ASD by constructing a gene-interaction network model32.
These examples highlight the progress made in artificial intelligence (AI) in the past few years, and its potential for healthcare applications in general and for ASD diagnostics in particular as the availability, diversity, and quality of relevant data grows, driven by the ability of ML models to find complex, non-linear relationships in the data compared to more traditional data analysis methods. The studies describe in detail the processes followed for the data processing and feature engineering steps. This is a key aspect of ML applications, as the data fed to the models is central to their performance.
Machine Learning Approaches
The quantity of data has a major impact as well. ML methods such as Support Vector Machines (SVM), Random Forest, Gradient Boosting and Deep Neural Network must be selected to suit the size of the dataset and type of data. Datasets with a large number of features per sample require more samples and more complex models, such as deep neural networks (DNN). These networks, with multiple layers of artificial neurons, or computational units, are capable of modelling non-linear relations, and are associated with the branch of AI called deep learning that has enabled recent breakthroughs in applications such as computer vision, speech recognition, language modelling and medical image analysis32-38.
In most of the ASD-related studies, a data-centric approach was adopted, where efforts focus on engineering available data to get the best result using classic ML algorithms33. This includes curating a subset of samples with pre-defined properties and then finding the subset of features that yield more robust predictions over the available dataset. Biomarker discovery studies use iterative approaches to develop effective ML models that obtain good diagnostic predictors28-30. Meta-analyses across multi-omics and microbiome studies have limitations in confounders such as sex-, age- and geography-related batch effects, compositionality, dimensionality, and sparsity. To address that, studies analyzing differential abundance of omics features have proposed algorithms such as Bayesian inference-based ranking algorithms as described in Morton et al. for the Gut-Brain Axis (GBA) disruption in ASD31. This necessity for data engineering reflects the need to get relevant results from datasets with limitations.
Given the importance of data and its limitations, synthetic data has emerged in recent years as a promising technology for ML applications. According to the Royal Society and The Alan Turing Institute: “Synthetic data is data that has been generated using a purpose-built mathematical model or algorithm, with the aim of solving a (set of) data science task(s)”39. Progress with DNN architectures, such as Generative Adversarial Networks, has extended the potential for generating data that reflects real-world data. Potential objectives for using synthetic data include patients’ privacy protection, data sharing facilitation, data augmentation and biases mitigation, all of which could be particularly relevant in healthcare applications40. While synthetic data offers great potential, it also presents major limitations that remain to be addressed and significant research is needed from a ML perspective and domain specific perspective. Critically, the quality of synthetic data is highly dependent on the quality and amount of the original data and the generation model.
Developing ML applications involves trade-offs between choice of models, quantity of data available and data engineering to improve the quality of the data. While there is not one definition of data quality, in the ML discipline we commonly refer to data that enables achievement of intended goals. While informative high-quality data can be hard to collect, behavioral data with limited quality and interpretation biases from hundreds or thousands of surveys is often more readily available. Abbas et al.22 studied 10 features in over 5,000 individuals with ASD and over 1,300 TD individuals. Feature engineering may be used to reduce interpretation bias, for example, in the development of diagnostic tools by focusing on finding a small subset of features with sufficient generalization power. In this respect, fMRI is conversely an extremely rich type of data, with large amount of information that is not necessarily relative to ASD itself and prone to noise41. Annotating this type of data and gathering large datasets is challenging and very time consuming. Authors can use pre-existing knowledge to select robust samples and to distill the data to correlations between Regions Of Interest (ROIs) so that it is possible to train a model on a smaller number of samples41.
Studies focusing on lower levels of systems biology such as transcriptomics, proteomics or metabolomics contain feature rich datasets with limited numbers of samples. These can be managed through data-centric methods to obtain potential diagnostic solutions28-30. However, the data sources are of higher quality and point to a broader range of questions that can be answered given sufficient resources and larger datasets. For example, Quillet et al.25 successfully used a pharmacometabolomics approach for distinguishing ASD groups and pharmacodynamics indications of cannabinoids using 645 features in 15 children with ASD and 9 TD children. This study linked metabolic changes in children with ASD to known biomarkers that can indicate clinical phenotype such as the stress biomarker cortisol and the aggression biomarker dehydroepiandrosterone sulfate (DHEA-S).
Bioinformatics Integration
Both supervised and unsupervised ML techniques have been successfully integrated with multi-omics databases. Feldner-Busztin et al.42 indicated the potential of the technologies while emphasizing the need to increase the sample size for each omics and the overall overlapping omics data per sample, namely the genomics, epigenomics, transcriptomics and metabolomics per sample. A range of analytical techniques are applied in several papers covering genomics32, RNA signature30,43, proteomics29, and metabolomics25,28. In particular, two high-level bioinformatics annotation engines (Gene Ontology: geneontology.org; and KEGG: www.genome.jp/kegg/) are applied across these patient-derived bioinformatics datasets to permit classification of genes, RNA, proteins and metabolites. Annotations and clusters demonstrate relevance: 1) to medical condition (in this case ASD vs. TD); 2) with cellular and organ location; 3) with metabolic pathways permitting elucidation of high-level effects such as inflammation; and 4) with neuronal activity (e.g. endocannabinoid pathways and neuronal signaling).
Future Perspective
We are at an inflection point where the omics and analytics fields are maturing, ML is being applied across omics data, and pharmacometabolomics biomarkers (cannabis-responsive) are being identified. Providing the right sample size and features with the available bioinformatics tissue-, patient-, cohort-, and pathophysiology-specific knowledge will allow ML applications to associate the current clinical phenotypes with underlying conditions of ASD and assist in diagnostic and therapeutic solutions. The growth in qualitative and quantitative data, the growing affordability of personal collecting devices and omics instruments together with standardization of databases show promise to provide the much-needed breakthroughs to effectively diagnose and treat ASD. These methods can also help to elucidate the extended endocannabinoid metabolism and related pathways, and to drive drug discovery and development, as well as to permit quantitative diagnosis for ASD.
Conclusions
Available ML techniques are sufficient to identify hidden interactions in large and complex datasets from individuals with ASD that link clinical phenotype to genotype, temporal changes in microbiome composition and medical cannabis treatment. The type of datasets currently used can answer many questions but there is a need for new data arising from experimental designs and detection tools specifically setup to answer some of the fundamental questions of ASD. Development of a sufficient quantity of informative high-quality, feature rich datasets that integrate omics, neuroimaging and bioinformatics in a dynamic mode such as before, during and after treatment will advance prediction and evaluation of treatment outcomes, and identification of the underlying conditions of ASD.
Abbreviations
ADI-R: Autism Diagnostic Interview-Revised
AI: Artificial intelligence
ADOS: Autism Diagnostic Observation Schedule
CBD: Cannabidiol
CBG: Cannabigerol
DBC: Developmental Behavior Checklist
fMRI: Functional Magnetic Resonance Imaging
ML: Machine Learning
THC: Tetrahydrocannabinol
TD: Typically developed
Conflict of Interest
I.K is co-founder and employee of Cannformatics, J.C.Q is consultant at Cannformatics and M.S.R is employee of Cannformatics. The authors declare that they are bound by confidentiality agreements that prevent them from disclosing their financial interests in this work.
References
- Huerta M, Lord C. Diagnostic evaluation of autism spectrum disorders. Pediatr Clin North Am. 2012; 59(1): 103-111.
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5. The American Psychiatric Association (APA): Arlington, VA, 2013.
- Lord C, Risi S, DiLavore PS, et al. Autism from 2 to 9 years of age. Arch Gen Psychiatry. 2006; 63(6): 694-670.
- Santos JF, Brosh N, Falk TH, et al. Very early detection of autism spectrum disorders based on acoustic analysis of pre-verbal vocalizations of 18-month old toddlers. 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. IEEE, 2013.
- Courchesne E, Pramparo T, Gazestani VH, et al. The ASD Living Biology: from cell proliferation to clinical phenotype. Mol psychiatry. 2019; 24(1): 88-107.
- Hornig M, Bresnahan MA, Che X, et al. Prenatal fever and autism risk. Mol Psychiatry. 2018; 23(3): 759-766.
- Parker W, Hornik CD, Bilbo S, et al. The role of oxidative stress, inflammation and acetaminophen exposure from birth to early childhood in the induction of autism. Int J Med Res. 2017; 45(2): 407-438.
- Becerra TA, Massolo ML, Yau VM, et al. A survey of parents with children on the autism spectrum: experience with services and treatments. Perm J. 2017; 21(2): 55-63.
- Dykens EM, Fisher MH, Taylor JL, et al. Reducing distress in mothers of children with autism and other disabilities: a randomized trial. Ped. 2014; 134(2): e454-e463.
- Al-Jawahiri R, Milne E. Resources available for autism research in the big data era: a systematic review. Peer J. 2017; 5: e2880.
- Washington P, Wall DP. A Review of and Roadmap for Data Science and Machine Learning for the Neuropsychiatric Phenotype of Autism. Annu Rev Biomed Data Sci. 2023; 6: 211-228.
- Keller S, Korkmaz G, Orr M, et al. The evolution of data quality: Understanding the transdisciplinary origins of data quality concepts and approaches. Annu Rev Stat Appl. 2017; 4(3): 85-108.
- Open Science Collaboration. Estimating the reproducibility of psychological science. Science. 2015; 349(6251): aac4716.
- Frye RE, Vassall S, Kaur G, et al. Emerging biomarkers in autism spectrum disorder: a systematic review. Ann Transl Med. 2019.
- Masini E, Loi E, Vega-Benedetti AF, et al. An overview of the main genetic, epigenetic and environmental factors involved in autism spectrum disorder focusing on synaptic activity. Int J Mol Sci. 2020; 21(21): 8290.
- Kang-Yi CD, Grinker RR, Beidas R, et al. Influence of community-level cultural beliefs about autism on families’ and professionals’ care for children. Transcult psychiatry. 2018; 55(5): 623-647.
- Yadav SP. The wholeness in suffix-omics,-omes, and the word om. JBT. 2007; 18(5): 277.
- Auslander N, Gussow AB, Koonin EV. Incorporating machine learning into established bioinformatics frameworks. Int J Mol Sci. 2021; 22(6): 2-19.
- Johnson CH, Gonzalez FJ. Challenges and opportunities of metabolomics. J Cell Physiol. 2012; 227(8): 2975-2981.
- Canario E, Chen D, Biswal B. A review of resting-state fMRI and its use to examine psychiatric disorders. Psychoradiology. 2021; 1(1): 42-53.
- Vabalas A, Gowen E, Poliakoff E, et al. Applying machine learning to kinematic and eye movement features of a movement imitation task to predict autism diagnosis. Sci Rep. 2020; 10(1): 8346-8361.
- Abbas H, Garberson F, Glover E, et al. Machine learning approach for early detection of autism by combining questionnaire and home video screening. JAMIA. 2018; 25(8): 1000-1007.
- Siani-Rose M, Cox S, Goldstein B, et al. Cannabis-responsive biomarkers: A pharmacometabolomics-based application to evaluate the impact of medical cannabis treatment on children with autism spectrum disorder. Cannabis Cannabinoid Res. 2023; 8(1): 126-137.
- Bent S, Lawton B, Warren T, et al. Identification of urinary metabolites that correlate with clinical improvements in children with autism treated with sulforaphane from broccoli. Mol Autism. 2018; 9: 1-12.
- Quillet JC, Siani-Rose, M, McKee R, et al. A machine learning approach for understanding the metabolomics response of children with autism spectrum disorder to medical cannabis treatment. Sci Rep. 2023; 13(1): 13022.
- Siani-Rose M, McKee R, Cox S, et al. The potential of salivary lipid-based cannabis-responsive biomarkers to evaluate medical cannabis treatment in children with autism spectrum disorder. Cannabis Cannabinoid Res. 2023; 8(4): 642-656.
- Hyde KK, Novack MN, LaHaye N, et al. Applications of supervised machine learning in autism spectrum disorder research: a review. Rev J Autism Dev Disord. 2019; 6: 128-146.
- West PR, Amaral DG, Bais P, et al. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS One. 2014; 9(11): e112445.
- Hewitson L, Mathews JA, Devlin M, et al. Blood biomarker discovery for autism spectrum disorder: A proteomic analysis. PLoS One. 2021; 16(2): e0246581.
- Voinsky I, Fridland OY, Aran A, et al. Machine learning-based blood RNA signature for diagnosis of autism spectrum disorder. Int J Mol Sci. 2023; 24(3): 2082.
- Morton JT, Jin DM, Mills RH, et al. Multi-level analysis of the gut–brain axis shows autism spectrum disorder-associated molecular and microbial profiles. Nat Neurosci. 2023; 26: 1208-1217.
- Krishnan A, Zhang R, Yao V. et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 2016; 19(11): 1454-1462.
- Cavus N, Lawan AA, Ibrahim Z, et al. A systematic literature review on the application of machine-learning models in behavioral assessment of autism spectrum disorder. J Pers Med. 2021; 11(4): 299.
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst. 2012; 25: 1-9.
- Chorowski JK, Bahdanau D, Serdyuk D, et al. Attention-based models for speech recognition. Adv Neural Inf Process Syst. 2015; 28: 1-9.
- Baevski A, Zhou Y, Mohamed A, et al. wav2vec 2.0: A framework for self-supervised learning of speech representations. Adv Neural Inf Process Syst. 2020; 33: 12449-12460.
- Brown T, Mann B, Ryder N, et al. Language models are few-shot learners. Adv Neural Inf Process Syst. 2020; 33: 1877-1901.
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017; 42: 60-88.
- Jordon J, Szpruch L, Houssiau F, et al. Synthetic Data - what, why and how. arXiv preprint 2023; arXiv:2205.03257.
- Giuffrè M, Shung DL. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. NPJ Digit Med. 2023; 6(1): 186.
- Chen CP, Keown CL, Jahedi A, et al. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage Clin. 2015; 8: 238-245.
- Feldner-Busztin D, Firbas Nisantzis P, Edmunds SJ, et al. Dealing with dimensionality: the application of machine learning to multi-omics data. Bioinformatics. 2023; 39(2): 1-8.
- Voinsky I, Zoabi Y, Shomron N, et al. Blood RNA Sequencing Indicates Upregulated BATF2 and LY6E and Downregulated ISG15 and MT2A Expression in Children with Autism Spectrum Disorder. Int J Mol Sci. 2022; 23(17): 1-14.
